One question that continues to remain unanswered during the ongoing coronavirus disease 2019 (COVID-19) pandemic has been why is there a distinctive hit-and-miss pattern when it comes to severe disease? An interesting new study that appears on the medRxiv* preprint server pinpoints one highly relevant risk factor: the state of the oral and gut microbiomes.
The spring wave of COVID-19 has filled many hospitals and intensive care units with patients gasping for breath. Many times that number are being screened for admission. The need for efficient and reliable risk biomarkers has never been greater.
Study details
The 4C mortality score was described in September 2020 to meet this need by the International Severe Acute Respiratory and Emerging Infections Consortium (ISARIC) and World Health Organization (WHO). This wide-ranging risk scoring system comprises eight variables, including age, sex, other pre-existing illnesses, level of consciousness, oxygen saturation in peripheral blood, and C-reactive protein (CRP).
With this information, however, the accuracy of prediction was only 79%, with 30% of patients at a high risk of mortality being missed. This drove the attempt to predict the risk of fatality by another method. The researchers in the current study made use of the fact that gut microbiomes in COVID-19 patients show severe disturbances, called dysbiosis, with 23 bacterial families being particularly linked to the severity of disease among patients who were hospitalized with COVID-19.
The scientists set up a robust framework using computational and analytical tools to trace the networks of links between the microbiota, clinical features, and severity of disease. They found that Enterococcus, a species of oral and intestinal bacteria, can robustly predict a fatal outcome in these patients.
This small study included 69 COVID-19 patients with moderate to severe symptoms, that is, those who required less than or more than 4 liters of oxygen, respectively. Of these, 63 had full medical records. Baseline clinical features were comparable in both groups, severe and moderate. Severe patients had to stay in hospital for six days more, on average, than moderately ill patients.
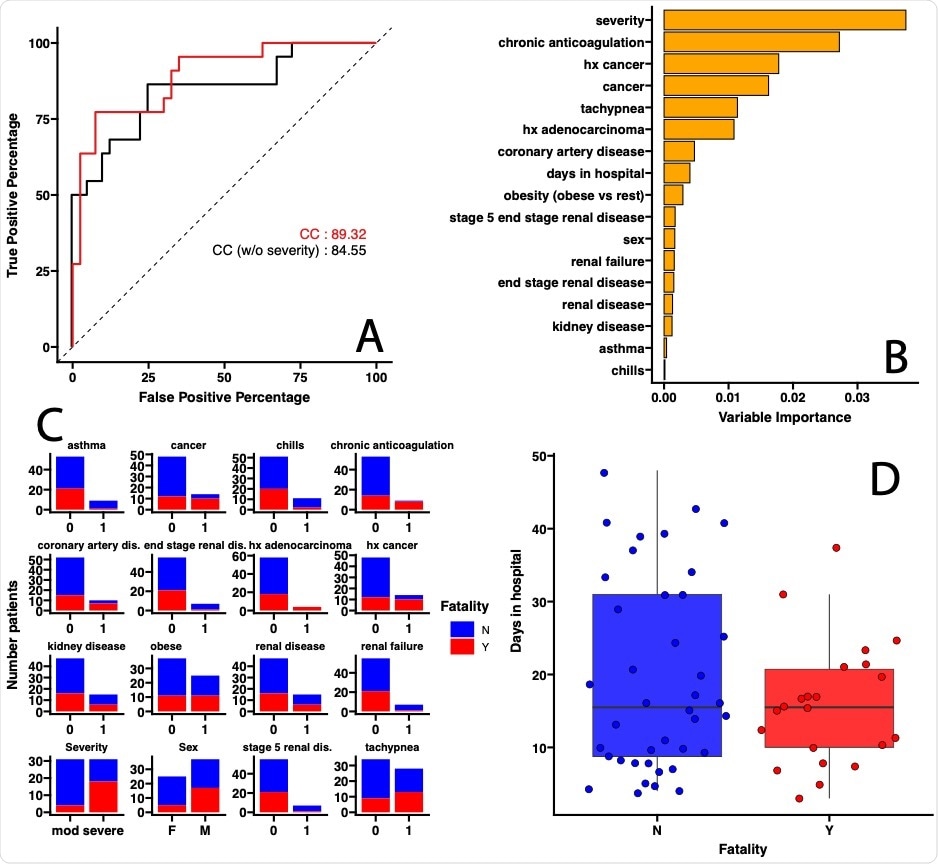
On analyzing the data on comorbidities, the researchers found that a combination of clinical variables, including the severity of COVID-19, had 89% accuracy in predicting a fatal outcome. In fact, the requirement for 4 liters of oxygen was the chief factor in predicting such an outcome. When the severity of disease was not considered, the accuracy dropped to 84%. This finding shows that respiratory symptoms are of importance in predicting COVID-19 outcomes.
Stool or oral microbiome predicts severity
It is known that a viral infection of the lung has a long-term impact on the gut microbiome. The researchers, therefore, made use of this knowledge to predict the severity of COVID-19, relating it to other common measures. They tested the effect of using clinical variables only, intestinal microbiome composition only, oral microbiome composition only, the first two combined, and the first and third in combination.
They found that the accuracy of the first model was ~76%. Again, the comorbidities that best predicted the severity of disease were those like high cholesterol, Latino race, coronary heart disease, asthma, obesity, breathing difficulty associated with hypoxia, rapid respiratory rate, number of days in hospital, thrombosis, and male sex.
Using the second and third models, with the microbiota of the stool or the mouth as predictors, they found accuracies of 92% and 84%, respectively. This is a 122% and 111% improvement in accuracy, respectively.
The combined models showed the highest prediction accuracy, at 96%, suggesting that the oral or gut microbiota are better at predicting COVID-19 severity. On further analysis of the microbiota, the researchers found an indicator species that can be cultured in the clinical laboratory.
Top predictor
The top three bacterial species for prediction of COVID-19 severity in the intestinal microbiome were Bacteroides uniformis, Enterococcus faecalis, and Monoglobus pectinilyticus, while those from the oral microbiome were Porphyromonas endodontalis, Veillonella tobetsuensis, and Bifidobacterium breve.
Directional analysis showed that a reduction in the abundance of Enterococcus faecalis, and Porphyromonas endodontalis, in the gut and mouth, respectively, in moderately sick COVID-19 patients, or a rise in the abundance of these pathological species in severely sick patients, were the best predictors of severe COVID-19.
Predictors of moderate COVID-19 included an increase in abundance of Bacteroides fragilis, Bacteroides caccae, and Clostridium clostridioforme, in the stool or Muribaculum intestinale in the mouth.
They could not detect any correlation between the number of bacteria of any species and antibody titers, even though higher anti-RBD IgG levels are correlated with survival. This may mean that the microbiota and IgG levels are independent predictors of severe outcomes.
Conclusion
“In this study, we have demonstrated that COVID-19 disease severity can be predicted by the stool or oral microbiome composition with higher accuracy than traditional clinical scoring methods. Particularly, two pathobionts in the either the oral (Porphyromonas endodontalis) or intestinal (Enterococcus faecalis) microbiota can serve as indicator species to robustly predict the severity of SARS-CoV-2 infections.”
This could lead to better risk stratification of patients, especially since Enterococcus faecalis is easy and inexpensive to culture. This could help provide earlier support for patients who are likely to develop lethal disease. The researchers urge that this bacterium be included in clinical risk stratification in the healthcare setting.
The severity of the disease is linked to uncontrolled inflammation, and this could be a result of gut dysbiosis, which has been incriminated in several chronic inflammatory conditions. This area requires further research, especially to understand the role of regulatory T cells (Tregs), which are responsible for immunomodulation in normal circumstances but can be abnormally expressed in COVID-19.
Such studies could help establish how “the dysbiosis in SARS-CoV-2 infected patients, and specifically the enrichment of the pathobionts we observed in this cohort, can contribute to COVID-19 disease severity via alteration of the Treg development.”
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.